Publications
Chromosomal instability and genomic alterations
in cholangiocarcinoma from Northeastern Thailand
Raksawan Deenonpoe, Molly A. Guscott,... Isabel McNeil... Nadeem Shaikh & Sarah E. McClelland
J Path 2025, DOI: 10.1002/path.6464
"This study provides increased understanding of the rate and potential mechanisms of CIN in CCA that may inform new therapeutic strategies that synergise with specific ongoing CIN mechanisms."

Experimental evolution of cancer chromosomal changes
Paper Review
Molly A. Guscott & Sarah E. McClelland
Nature Genetics 2024, DOI: 10.1038/s41588-024-01742-6
"Experimental genome evolution of normal human cells reveals an intrinsic propensity to develop cancer-associated chromosomal alterations."

Targeted assembly of ectopic kinetochores to induce chromosome-specific segmental aneuploidies
Laura Tovini & Sarah C Johnson & Sarah E. McClelland
EMBO 2023, DOI: 10.15252/embj.2022111587
"Our findings provide new insights into ectopic kinetochore biology and represent an important step towards investigating the role of specific aneuploidy and chromosome mis-segregation events in diseases associated with aneuploidy."

Literature Review
Disentangling the roles of aneuploidy, chromosomal instability and tumour heterogeneity in developing resistance to cancer therapies
Joana Reis Andrade, Annie Dinky Gallagher, Jovanna Maharaj & Sarah E. McClelland

Chromosome Research 2023, DOI: 10.1007/s10577-023-09737-5
"Aneuploidy, CIN and ITH have been associated with poor prognosis in cancer, and a wealth of evidence suggests they contribute, either alone or in combination, to cancer therapy resistance. A full understanding of the interplay between aneuploidy, CIN and ITH is required to tackle therapy resistance in cancer patients."
Replication stress generates distinctive landscapes of DNA copy number alterations and chromosome scale losses
Nadeem Shaikh, Alice Mazzagati, Simone De Angelis, Sarah C. Johnson... & Sarah E. McClelland
Genom Biology 2022, DOI: 10.1186/s13059-022-02781-0
"Motivated by a desire to understand the mechanisms that convert replication stress to genome evolution during cancer, we sought to comprehensively assess the impact that replication stress has on the genome. Using a single-cell sequencing approach, we were able to detect CNAs caused by replication stress within a single-cell cycle and in the absence of any selective pressure, providing a comprehensive and unbiased analysis of genomic instability caused by two distinct classes of replication stress."

Literature Review
The multifaceted role of micronuclei in tumour progression: A whole organism perspective.
Molly A. Guscott, Akash Saha, Jovanna Maharaj & Sarah E. McClelland
iJBCB 2022, DOI: 10.1016/j.biocel.2022.106300
"We discuss the further consequences of micronucleus formation, including how structural changes to the micronuclear envelope, and the rupturing of micronuclear membranes can contribute to metastasis, immune cell activation and overall, tumour progression."

Diversity in chromosome numbers promotes resistance to chemotherapeutics.
Paper Review
Nadeem Shaik & Sarah E. McClelland
DevCell 2021, DOI: 10.1016/j.devcel.2021.08.017
"In this issue of Development Cell, we discuss the publications by from Ippolito et al. and from Lukow et al. these show that increasing the range of aneuploidy states in cells increases their chance of developing resistance when they are subjected to chemotherapy."
Specific Mechanisms of Chromosomal Instability Indicate Therapeutic Sensitivities in High-Grade Serous Ovarian Carcinoma
Naoka Tamura, Nadeem Shaikh, Daniel Muliaditan, Tanya N. Soliman... & Sarah E. McClelland
AACR 2021, DOI: 10.1158/0008-5472.CAN-19-0852
"We show that HGSC CIN is complex and suggest that specific CIN mechanisms could be used as functional biomarkers to indicate appropriate therapy."

Paper Review
Watching cancer cells evolve through chromosomal instability
Sarah C. Johnson & Sarah E. McClelland
Nature 2019, DOI: 10.1038/d41586-019-01709-2
"Chromosomal abnormalities are a hallmark of many types of human cancer, but it has been difficult to observe such changes in living cells and to study how they arise. Progress is now being made on this front."

The emerging links between chromosomal instability (CIN), metastasis, inflammation and tumour immunity
Literature Review
Andréa E. Tijhuis, Sarah C. Johnson & Sarah E. McClelland
Molecular Cytogenetics 2019, DOI: 10.1186/s13039-019-0429-1
"This review is focused on the investigation of possible links between CIN, metastasis and the host immune system in cancer development and treatment. We specifically focus on these links since most cancer deaths are due to the consequences of metastasis, and immunotherapy is a rapidly expanding novel avenue of cancer therapy."

Impaired CENP-E Function Renders Large Chromosomes More Vulnerable to Congression Failure
Laura Tovini & Sarah E. McClelland
Biomolecules 2019, DOI: 10.3390/biom9020044
"We observed a bias in congression efficiency related to chromosome size, with larger chromosomes more sensitive to CENP-E inhibition. This bias is likely due to two contributing factors; an initial propensity of larger chromosomes to be peripheral and thus rely more upon CENP-E function to migrate to the metaphase plate, and additionally a bias between specific chromosomes' ability to congress from a polar state."

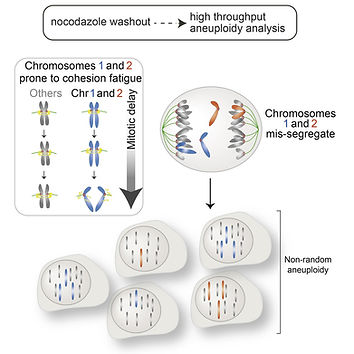
Non-random Mis-segregation of Human Chromosomes
Joseph T. Worrall, Naoka Tamura, Alice Mazzagatti, Nadeem Shaikh... & Sarah E. Mclelland
Cell Reports 2018, DOI: 10.1016/j.celrep.2018.05.047
"We show that aneuploidy occurs non-randomly following common treatments to elevate chromosome mis-segregation. Temporary spindle disruption leads to elevated mis-segregation and aneuploidy of a subset of chromosomes, particularly affecting chromosomes 1 and 2. Unexpectedly, we find that a period of mitotic delay weakens centromeric cohesion and promotes chromosome mis-segregation and that chromosomes 1 and 2 are particularly prone to suffer cohesion fatigue."